In order to make our curation data machine-readable and thus enhance communication with other biodiversity resources on the web for knowledge sharing and data integration, we create semantic web data described below:
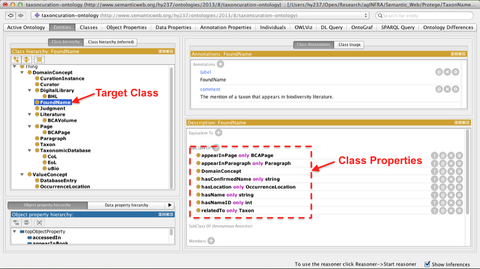
Figure 1. An OWL ontology for taxonomic name curation shown in the Protégé tool
1. A Light-weight OWL Ontology for Taxon Name Curation
One of the primary benefits to build an ontology is to allow modeling vocabulary and semantics within a domain of knowledge. We create a light-weight OWL ontology to support literature-based curation for taxonomic name databases using the Protégé Ontology tool. This OWL ontology is used to add semantic meaning into biodiversity data within RDF graph models (discussed later). As shown in Figure 1, the classes/subclasses defined in the ontology can be grouped into two main categories:
(a) DomainConcept
This category lists all the domain-specific classes used for taxon name curation, which include text-related concepts (e.g., BookVolume, BookPage, Paragraph, and FoundName), biodiversity-related concepts (e.g., Taxon, CoL, EoL, uBio, BHL) and validation-related concepts (e.g. CurationInstance, Judgment, Curator).
(b) ValueConcept
ValueConcept contains the classes (e.g., DatabaseEntry and OccurrenceLocation) which are used to describe only values of the classes or subclasses from DomainConcept.
OWL Ontology Resource
The OWL ontology for literature-based taxon name curation, TaxonCuration_Ontology.owl, is available here [download]
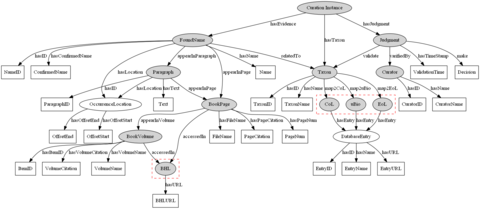
Figure 2. A RDF graph data model for taxonomic name curation [Picture]
2. RDF Graph Databases
We store the curation data into a semantic data model called RDF Graph database. In the RDF graph data model (see Figure 2), the semantic interactions between curation resources are encoded with our created OWL ontology discussed above. Each node in the graph represents either a resource or a liberal. A resource is a object with related properties (attributes), e.g., the BookVolume concept, while a liberal refers to a data value, e.g., a TaxonName string - "Pisces". In the graph model, the nodes with the eclipse shape are resources, and the ones with the box shape are literals. The shade resources refer to the classes in the DomainConcept category of the ontology, whereas the blank resources are the ones in the ValueConcept category. The resources contained in the broken red box are the existing biodiversity resources available on the web.
![]()
Figure 3. A simple data graph
A RDF graph database contains a collection of RDF statements known as RDF Triples. Each RDF statement consists of three parts: subject (resource identifier), predicate (property name), and object (property value). Figure 3 shows a simple data graph that can be depicted with two RDF statements: (1) Subject [BookPage] - Predicate [appearInVolume] - Object [BookVolume] (2) Subject [BookVolume] - Predicate [hasItemID] - Object [ItemID: "1321"]. More detains about RDF statement can be found in the W3 School tutorial.
RDF Semantic Data
The semantic web data that we created using Apache Jena contains the curation data about 4 digitized Biologia Centrali-Americana (BCA) volumes. They are:
- Bird BCA Volume: Biologia Centrali-Americana, Aves, Volume I (1879-1904) by Osbert Salvin and F. DuCane Godman. [RDF File]
- Fish BCA Volume: Biologia Centrali-Americana, Pisces (1906-1908) by Charles Tate Regan. [RDF File]
- Beetle I BCA Volume: Biologia Centrali-Americana, Insecta, Coleoptera, Volume I, Part 1 (1881-1884) by Henry Walter Bates. [RDF File]
- Beetle II BCA Volume: Biologia Centrali-Americana, Insecta, Coleopetera, Rhynchophora, Volume IV, Part 3 (1889-1911) by David Sharp and G. C. Champion. [RDF File]
We also create a RDF dataset that contains the whole curation data about these 4 BCA volumes, RDF_TaxonCuration.rdf. [RDF File]
3. Access Semantic Data Using SPARQL Queries
SPARQL is a Web Query Language that is used to query data conforming to the RDF data model. The syntax of a SPARQL query is typically a triple pattern, which is similar to a RDF triple, but may includes variables to add flexibility in how it matches against the data. Figure 4 shows a SPAQRL Query example for the search of curation evidence from a BCA volume ("mobotbca_03_01_00") given a taxonomic name("Amblycercus holosericeus").
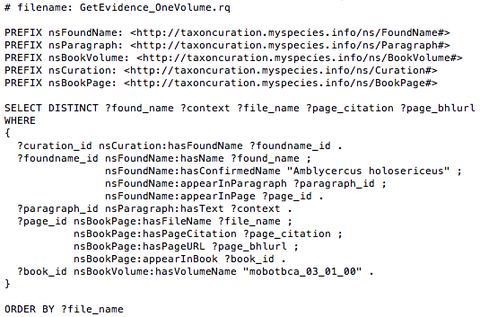
Figure 4. A SPARQL Query example to access the RDF semantic data about the curation information
There are two ways to access our curated RDF semantic data using SPARQL queries:
(a) Jena AQR query engine for local RDF data
Jena AQR is a query engine for Jena that supports the SPARQL RDF query language. ARQ includes a batch file about SPARQL query statements and a command-line shell script, which allow the user to run the GetEvidence_OneVolume.rq query against the RDF_TaxonCuration.rdf data as shown below:
![]()
(b) Jena Fuseki server over HTTP
Method 1: Jena Fuseki is a SPARQL Endpoint server, which provides HTTP interface to access the RDF data. Jena Fuseki service offers SPARQL Query, SPARQL Update and SPARQL Graph Store protocol, as well as file upload, on an in-memory dataset. You can enter SPARQL query statements in the SPARQL Query panel of the Fuseki Query form as shown in Figure 5, and retrieve the information of interest by clicking the Get Results button.
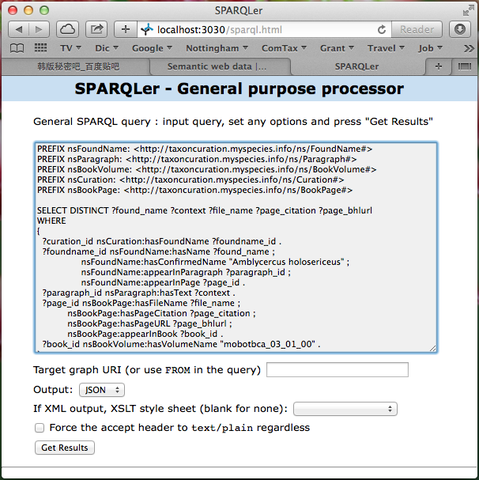
Figure 5. SPARQL Endpoint provided by Jena Fuseki server
Method 2: Another way to access the RDF data is to use the script control provided by Jena Fuseki Server through SOH commands. SOH (SPARQL Over HTTP) is a set of command-line scripts for working with SPARQL 1.1. SOH is server-independent and will work with any compliant SPARQL 1.1 system offering HTTP access. You can obtain the relevant curation information by executing the following SOH commands:
![]()
![]()
